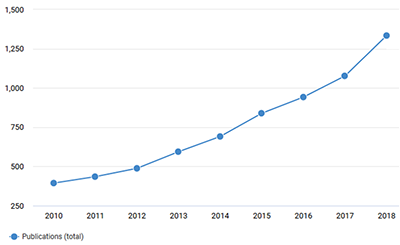
The use of environmental DNA (eDNA) is gaining in popularity and profile, partly driven by technological advances, partly by the urgent requirement for measuring natural habits for indigenous and alien species using diagnostic tools and the steeply rising importance of such measurements for environmental stewardship. This is mirrored by the rapid rise in scientific publications citing “environmental DNA” (see figure), and the recent launch of a scientific journal Environmental DNA.
But what exactly is “environmental DNA” that makes it uniquely different from other DNA samples?
Unfortunately, there is no clear answer, but this lack of clarity opens the risk that some scientists may miss opportunities to bring their work to a wider audience, and environmental practitioners and policy makers may miss relevant scientific knowledge and developments.
Increasing numbers of scientific papers use the term "environmental DNA"
If we take one of the definitions of eDNA from the scientific literature, it is “genetic material obtained directly from environmental samples (soil, sediment, water, etc.) without any obvious signs of biological source material” (Thomsen et al 2015), a definition which hinges on whether you can see (and identify) the source of the DNA. Under this definition there are both the ‘classic’ examples, such as the eDNA test for great crested newts (where pond water samples were sent into a lab and the water tested for newt DNA), and environmental samples of microorganisms, such as identifying the bacteria in a sample of soil or water using their DNA. A problem with this definition of eDNA is that it includes well-established fields of scientific research, where the scientists may not use this term to describe what they are doing. As an example, metabarcoding DNA from soil to identify the bacterial and fungal communities fits this definition, as the biological source material (the individual bacterial or fungal species) are not obviously present to an observer. However, of around 3500 articles on soil metabarcoding in Google Scholar since 2015, only 1300 of those also contain the term eDNA. The researchers in this area would not necessarily consider themselves to be studying “eDNA”. More stark is the difference in aerial spore trapping; again, taking samples from the air to collect spores and identifying the species present using DNA fits the definition of eDNA, but of the around 1800 articles since 2015 on “aerial spore trapping DNA”, only 69 of those also contain the term eDNA. The scientists working in this area clearly don’t find this definition of eDNA relevant to them, and don’t use it. Many of the topics subsumed into this rather catch-all definition of “eDNA” are already well established areas of research, and the expectation that this research will be described under the heading “eDNA” means that this large background knowledge could be overlooked. It also means that scientists may not be aware of forums they can usefully contribute to: if a working group is formed to discuss using DNA to monitor the environment but is referred to as an “eDNA” forum, then some scientific experts in the field may not appreciate that they can bring their knowledge to bear.


An alternative definition, the spirit of which is gaining in popularity among many scientists in the eDNA field, is summarised on Wikipedia as: “environmental DNA or eDNA is DNA that is collected from a variety of environmental samples such as soil, seawater, snow or even air rather than directly sampled from an individual organism”. The distinction here is that the DNA is not from organisms themselves but is DNA that has been shed into the environment (skin cells, gametes, faeces) by the organisms. This, therefore, excludes studies where the organisms are extracted from a sample and blended up to release their DNA. This definition is more robust to some of the problems outlined above and has the benefit that it is distinctly different to already well established fields. There is still some ambiguity whether micro-organisms’ bulk DNA-extracted from a sample counts as eDNA or not but, on the whole, this seems a less problematic definition.
Settling on this more restrictive definition would seem a way forward, should the scientific community concur. However, in discussions with some environmental practitioners and policy makers, I often find that eDNA is being used with an alternative meaning of the use of DNA-based methods to monitor the environment. In this case, the ‘environment’ in ‘environmental DNA’ describes the use the DNA is being put to, rather than the type of sample it is. An example of this alternative use is describing the identification of insects from a pitfall trap as an environmental DNA method; using DNA from the organisms themselves comes firmly back into scope, and soil and fungal community analysis from soil is unambiguously within this definition. This approximate definition is in direct conflict with the definition that sections of scientific community is adopting, but it is in no way helpful to free-flowing discussions to pedantically mutter: “Well, I think you’ll find that’s not actually eDNA”. It is also a definition that seems to have been pragmatically adopted the recently launched journal Environmental DNA, which lists “[…] ancient DNA, non-invasive sampling, diet analyses, metabarcoding, metagenomics, microbial ecology and pathogens” as being in scope (Bernatchez 2019). Although eDNA is a popular area of study, it is dwarfed by the size of the metabarcoding scientific community, and the eDNA community (scientists, managers, policy makers) only stands to lose if it does not actively linking to the metabarcoding community.
As Fera is a translational science organisation, and all about discussions between scientists and practitioners, how do we use the term? In general, we use the term eDNA in the strict sense of free DNA captured from the environment, and avoid the problems around the broader definition by using phrases such as “DNA to monitor the environment”. In the absence of the perfect lexicon, I believe we can live with this ambiguity as long as we understand that when we discuss eDNA, we may not all using the same definition.
- Eleanor Jones




